%matplotlib inline
We will be covering
- Marking bad channels
- Marking bad epochs
- Using autoreject to automatically mark bad channels and epochs
Let’s first load the raw data as usual
import numpy as np
import mne
from mne.datasets import sample
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True) # already has an EEG ref
Opening raw data file /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Marking bad channels
If you already know which are the bad channels, simply do:
raw.info['bads'] = ['MEG 2443']
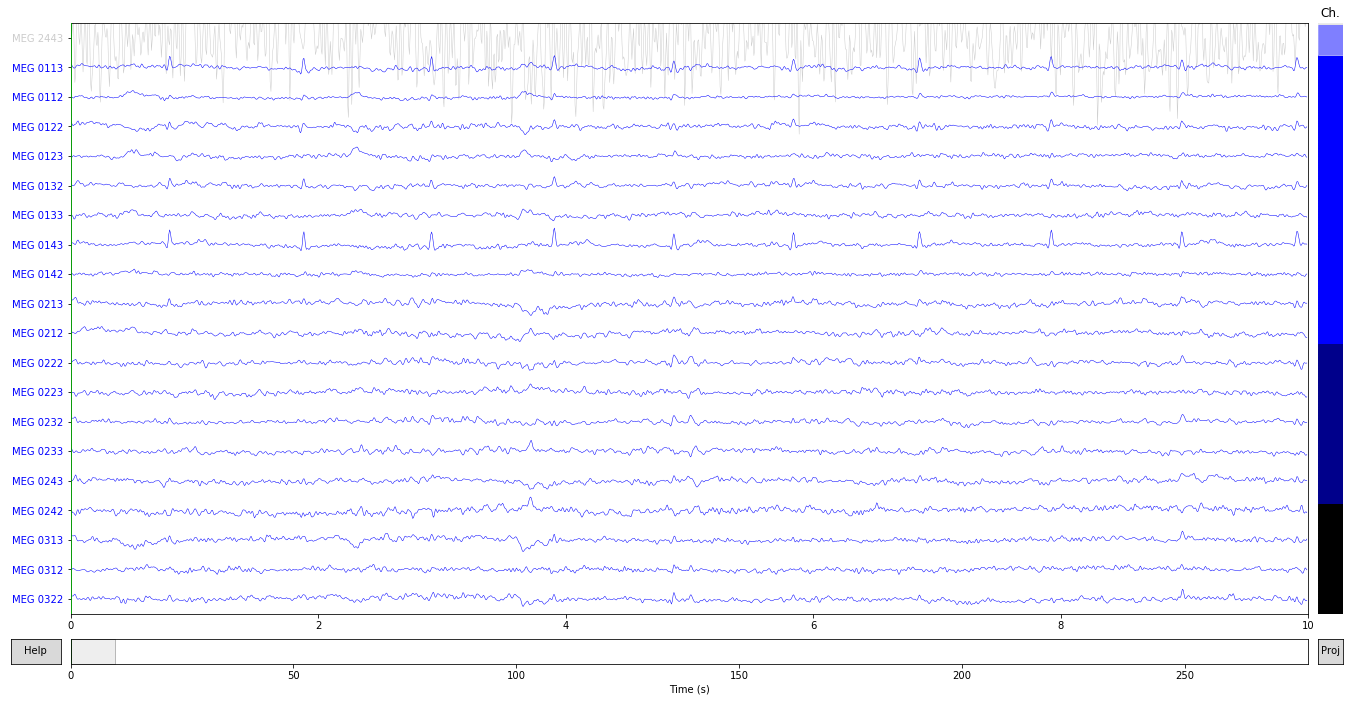
You can also mark them interactively in raw.plot(). The bad channel is shown in gray
ch_names = raw.info['ch_names'].copy()
ch_names.remove('MEG 2443')
raw.reorder_channels(['MEG 2443'] + ch_names)
raw.plot();
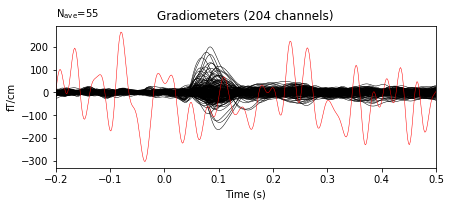
It’s not only raw. You can mark bads also in evoked. Let’s first read in the evoked data.
# Reading data with a bad channel marked as bad:
fname = data_path + '/MEG/sample/sample_audvis-ave.fif'
evoked = mne.read_evokeds(fname, condition='Left Auditory',
baseline=(None, 0))
evoked.pick_types(meg='grad', eeg=False, exclude=[])
# plot with bads
evoked.plot(exclude=[], time_unit='s');
Reading /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
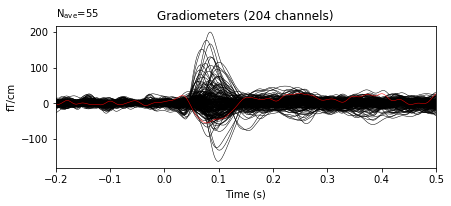
It’s also possible to repair the bad channels using interpolation
evoked.interpolate_bads(reset_bads=False, verbose=False);
Let’s plot the cleaned data
evoked.plot(exclude=[], time_unit='s');
Marking bad epochs
MNE allows you to specify rejection dictionary based on peak-to-peak thresholds for each channel type
reject = dict(grad=4000e-13, mag=4e-12, eog=200e-6)
events = mne.find_events(raw, stim_channel='STI 014')
event_id = {"auditory/left": 1}
tmin, tmax = -0.2, 0.5
baseline = (None, 0) # means from the first instant to t = 0
picks = mne.pick_types(raw.info, meg=True, eeg=True, eog=True,
stim=False, exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True,
picks=picks, baseline=baseline, reject=reject,
preload=True)
319 events found
Event IDs: [ 1 2 3 4 5 32]
72 matching events found
Applying baseline correction (mode: mean)
Not setting metadata
Created an SSP operator (subspace dimension = 4)
4 projection items activated
Loading data for 72 events and 106 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on MAG : ['MEG 1711']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
9 bad epochs dropped
You can also reject after constructing epochs, just do:
reject.update({'eog': 150e-6})
epochs.drop_bad(reject=reject)
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
8 bad epochs dropped
<Epochs | 55 events (all good), -0.199795 - 0.499488 sec, baseline [None, 0], ~19.9 MB, data loaded,
'auditory/left': 55>
But the thresholds need to be stricter each time.
Tuning rejection thresholds
import matplotlib.pyplot as plt
from ipywidgets import interact
picks = mne.pick_types(raw.info, meg=False, eeg=True)
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True,
picks=picks, baseline=baseline, reject=None,
preload=True)
def reject_epochs(reject):
reject = dict(eeg=reject * 1e-6)
evoked = epochs.copy().drop_bad(reject=reject, verbose=False).average()
evoked.plot(spatial_colors=True)
print('Number of epochs retained: %d/%d' % (evoked.nave, len(epochs)))
72 matching events found
Applying baseline correction (mode: mean)
Not setting metadata
Created an SSP operator (subspace dimension = 1)
4 projection items activated
Loading data for 72 events and 106 original time points ...
0 bad epochs dropped
interact(reject_epochs, reject=(35, 250, 10));
interactive(children=(IntSlider(value=135, description='reject', max=250, min=35, step=10), Output()), _dom_cl…
Autoreject
Autoreject (global) can compute the rejection dictionary automatically
from autoreject import get_rejection_threshold # noqa
reject = get_rejection_threshold(epochs)
print(reject)
Estimating rejection dictionary for eeg
{'eeg': 8.687666582562178e-05}
Autoreject (local) finds per channel thresholds:

import numpy as np
from autoreject import AutoReject
n_interpolates = np.array([1, 2, 4])
consensus = np.linspace(0.5, 1.0, 6)
ar = AutoReject(n_interpolates, consensus, thresh_method='random_search',
random_state=42)
Now, we find the rejection thresholds per-channel and optimize for the number of channels to be interpolated.
raw.info['bads'] = []
picks = mne.pick_types(raw.info, meg='grad', eeg=False)
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=True,
picks=picks, baseline=baseline, reject=None,
preload=True)
# Note that fitting and transforming can be done on different compatible
# portions of data if needed.
ar.fit(epochs['auditory/left'])
72 matching events found
Applying baseline correction (mode: mean)
Not setting metadata
4 projection items activated
Loading data for 72 events and 106 original time points ...
0 bad epochs dropped
84 coil definitions read
Running autoreject on ch_type=grad
[............................................................] 100.00% Creating augmented epochs |
[............................................................] 100.00% Computing thresholds ... |
[............................................................] 100.00% Repairing epochs |
[ ] 0.00% n_interp |
[............................................................] 100.00% Repairing epochs |
[.................... ] 33.33% n_interp /
[........................................................... ] 98.61% Repairing epochs \
Now, we can look at the rejection thresholds for each channel
for ch_name in epochs.info['ch_names'][:5]:
print('%s: %s' % (ch_name, ar.threshes_[ch_name]))
# plt.hist(np.array(list(ar.threshes_.values())), 30, color='g', alpha=0.4)
We can check what applying autoreject would do to the epochs:
- Good data (light red)
- Bad segment but not to be interpolated (medium dark red)
- Bad segment to be interpolated (dark red)
reject_log = ar.get_reject_log(epochs['auditory/left'])
reject_log.plot()
Another way to visualize this is to plot them on the epochs
reject_log.plot_epochs(epochs['auditory/left']);
We can apply these rejection thresholds to new (or to the old) data:
epochs_clean = ar.transform(epochs['auditory/left'])
evoked = epochs['auditory/left'].average()
evoked.info['bads'] = ['MEG 2443']
evoked.plot(exclude=[]);
evoked_clean = epochs_clean.average()
evoked_clean.info['bads'] = ['MEG 2443']
evoked_clean.plot(exclude=[]);
For more info, visit