%matplotlib inline
Signal Space Projection (SSP) Math
We can write the measured signal $b \in \mathbb{R}^{C \times T}$ as
where $b_s\in \mathbb{R}^{C \times T}$ is the brain signals and $b_n(t) \in \mathbb{R}^{C \times T}$ is the noise
Noise $b_n$ can be:
- ambient sensor noise
- electrophysiological corruption
- cardiac
- ocular
Now, we can apply PCA on $b_n$ and keep only top $M$ components with loadings $ c_n \in \mathbb{R}^M$:
where $U = [b_1, …, b_M] \in \mathbb{R}^{C \times M}$ are the orthonormal basis.
Now, the SSP operator $P_{\perp} \in \mathbb{R}^{C \times C}$ is given by:
so that:
Artifact Correction with SSP
This tutorial explains how to estimate Signal Subspace Projectors (SSP) for correction of ECG and EOG artifacts.
import numpy as np
import mne
from mne.datasets import sample
from mne.preprocessing import compute_proj_ecg, compute_proj_eog
# getting some data ready
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
raw = mne.io.read_raw_fif(raw_fname, preload=True)
Opening raw data file /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
We can see that the raw data has projectors computed but not yet applied.
print(raw.info['projs'])
[<Projection | PCA-v1, active : False, n_channels : 102>, <Projection | PCA-v2, active : False, n_channels : 102>, <Projection | PCA-v3, active : False, n_channels : 102>, <Projection | Average EEG reference, active : False, n_channels : 60>]
Exercise
Where did these projectors come from? What was the noise $b_n$?
To interactively explore the effect of applying proj, use raw.plot() and click on the “proj” button
# raw.plot()
Now, we can apply the projectors by doing:
raw.apply_proj()
print(raw.info['projs'])
Created an SSP operator (subspace dimension = 4)
4 projection items activated
SSP projectors applied...
[<Projection | PCA-v1, active : True, n_channels : 102>, <Projection | PCA-v2, active : True, n_channels : 102>, <Projection | PCA-v3, active : True, n_channels : 102>, <Projection | Average EEG reference, active : True, n_channels : 60>]
Compute SSP projections
First let’s do ECG.
projs, events = compute_proj_ecg(raw, n_grad=1, n_mag=1, n_eeg=0, average=True, verbose=False)
print(projs)
ecg_projs = projs[-2:]
mne.viz.plot_projs_topomap(ecg_projs);
[<Projection | PCA-v1, active : True, n_channels : 102>, <Projection | PCA-v2, active : True, n_channels : 102>, <Projection | PCA-v3, active : True, n_channels : 102>, <Projection | Average EEG reference, active : True, n_channels : 60>, <Projection | ECG-planar--0.200-0.400-PCA-01, active : False, n_channels : 203>, <Projection | ECG-axial--0.200-0.400-PCA-01, active : False, n_channels : 102>]
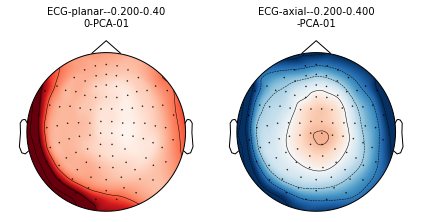
Now let’s do EOG. Here we compute an EEG projector, and need to pass the measurement info so the topomap coordinates can be created.
projs, events = compute_proj_eog(raw, n_grad=1, n_mag=1, n_eeg=1, average=True, verbose=False)
print(projs)
eog_projs = projs[-3:]
mne.viz.plot_projs_topomap(eog_projs, info=raw.info);
[<Projection | PCA-v1, active : True, n_channels : 102>, <Projection | PCA-v2, active : True, n_channels : 102>, <Projection | PCA-v3, active : True, n_channels : 102>, <Projection | Average EEG reference, active : True, n_channels : 60>, <Projection | EOG-planar--0.200-0.200-PCA-01, active : False, n_channels : 203>, <Projection | EOG-axial--0.200-0.200-PCA-01, active : False, n_channels : 102>, <Projection | EOG-eeg--0.200-0.200-PCA-01, active : False, n_channels : 59>]
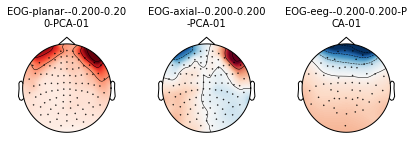
Exercise
What are we plotting here? Is it the SSP operator?
Add SSP projections
MNE is handling projections at the level of the info, so to register them populate the list that you find in the ‘proj’ field
raw.info['projs'] += eog_projs + ecg_projs
# raw.add_proj
Now MNE will apply the projs on demand at any later stage,
so watch out for proj parmeters in functions or to it explicitly
with the .apply_proj method
Demonstrate SSP cleaning on some evoked data
events = mne.find_events(raw, stim_channel='STI 014')
reject = dict(grad=4000e-13, mag=4e-12, eog=150e-6)
# this can be highly data dependent
event_id = {'auditory/right': 2}
epochs_no_proj = mne.Epochs(raw, events, event_id, tmin=-0.2, tmax=0.5,
proj=False, baseline=(None, 0), reject=reject,
verbose=False)
epochs_no_proj.average().plot(spatial_colors=True, time_unit='s');
epochs_proj = mne.Epochs(raw, events, event_id, tmin=-0.2, tmax=0.5, proj=True,
baseline=(None, 0), reject=reject, verbose=False)
epochs_proj.average().plot(spatial_colors=True, time_unit='s');
319 events found
Event IDs: [ 1 2 3 4 5 32]
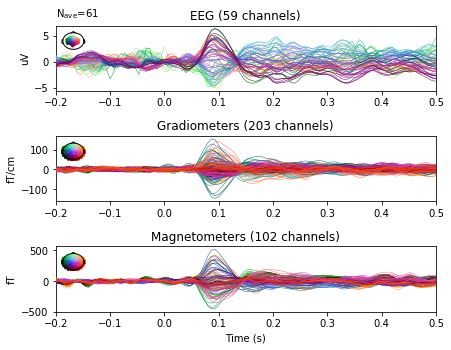
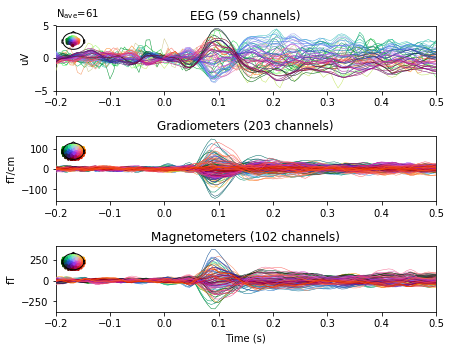
Interactive topomaps with SSP applied
raw = mne.io.read_raw_fif(raw_fname, preload=True)
evoked = mne.Epochs(raw, events, event_id, tmin=-0.2, tmax=0.5,
proj='delayed', baseline=(None, 0),
reject=reject, verbose=False).average()
# set time instants in seconds (from 50 to 150ms in a step of 10ms)
times = np.arange(0.05, 0.15, 0.01)
fig = evoked.plot_topomap(times, proj='interactive', time_unit='s')
Opening raw data file /home/mainak/Desktop/projects/github_repos/mne-python/examples/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Current compensation grade : 0
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
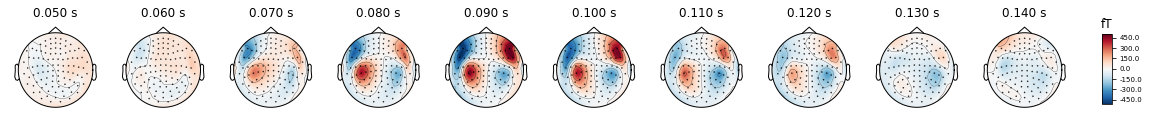

Now you should see checkboxes. Remove a few SSP and see how the auditory pattern suddenly drops off
Exercises
1) Can you compute your own proj using empty room data?
erm_fname = data_path + 'sample/MEG/ernoise_raw.fif'
mne.compute_proj_raw?
2) How would you compute the SSP from the evoked baseline instead of empty room?
raw = mne.io.read_raw_fif(raw_fname, preload=True, verbose=False)
reject = dict(grad=4000e-13, mag=4e-12, eog=150e-6)
evoked = mne.Epochs(raw, events, event_id, tmin=-0.2, tmax=0.5,
proj='delayed', baseline=(None, 0),
reject=reject, verbose=False).average()
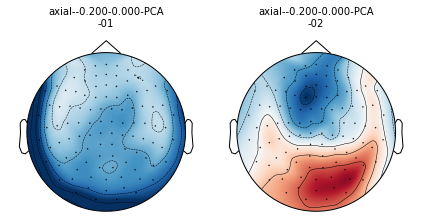
projs = mne.compute_proj_evoked(evoked.copy().crop(tmax=0), n_grad=0, n_mag=2, n_eeg=0)
mne.viz.plot_projs_topomap(projs, info=evoked.info);
Adding projection: axial--0.200-0.000-PCA-01
Adding projection: axial--0.200-0.000-PCA-02
Try making reject=None. What happens? Why?